Metabolism
Laws of Thermodynamics
1) The total amount of energy in the universe is constant. Energy is neither created nor destroyed but converted from one form to another. A gain in energy in an object or process means a loss of energy elsewhere in the universe.
2) The entropy of the universe increases with any change that occurs. Things become more random and disorganized.
-Catabolic reaction; reaction that breakdown complex substances
-Anabolic reaction; reaction that builds complex substances from simpler subunits
-Collectively, the sum of all anabolic and catabolic processes in a cell or organism is called metabolism
-Energy is absorbed when reactant bonds break and energy us released when product bonds form
-Activation energy is the amount of energy needed to strain and break the reactants' bond
-Entropy; a measure of the randomness or disorder in energy or in a collection of objects
-Free energy; energy that can do useful work
-Organisms have cells that go through anabolic processes that seems to violate the second law of thermodynamics by apparently causing the universe to become a little more ordered
-Order by anabolic processes is overshadowed by greater disorder from catabolic processes.
-Living organisms build up their bodies and the world around them at the expense of the entropy of the universe as a whole
-Exergonic reaction; spontaneous with a decrease in free energy
-Endergonic reaction; not spontaneous with an increase in free energy
-Adenosine triphosphate (ATP) is the primary source of free energy in living cells. When a phosphate is broken from the chain, free energy is released
-Phosphorylation is the process of attaching a phosphate group to an organic molecule such as ADP which causes the molecule to become more reactive
Enzymes
-Enzymes are protein catalysts that speeds up a chemical reaction without being consumed in the processed
-Competitive inhibitors can block a normal substrate from binding to an enzyme's active site
-Noncompetitive inhibitors alter the enzyme's shape so it loses it's affinity for its substrate
-Allosteric sites on the enzymes can have substances bind to it that affect the active sites of the enzyme
-Feedback inhibition is a method of metabolic control in which a product formed later in a sequence of reactions allosterically inhibits an enzyme that catalyzes a reaction occurring earlier in the process
Thursday, October 20, 2011
Tuesday, October 18, 2011
Blog 4: 10 Points to Know About Biotechnology
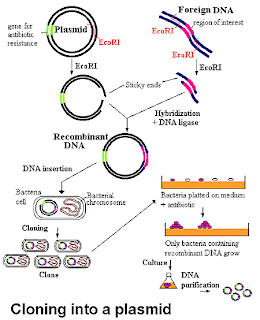
Vector Cloning
-Vector cloning requires plasmids, restriction enzymes, ligase, and a vector (bacteria).
-"Sticky ends" are single strand sequences at the ends of the cut pieces that matches with other sticky ends made by the same restriction enzyme.
Polymerase Chain Reaction
-DNA that has been treated by restriction enzymes can be sorted into their respective lengths.
-Smaller fragments move through the gel to the positively-charged terminal (DNA has a negative charge) while larger fragments remain closer to the starting point.
DNA Sequencing
-DNA is replicated in solutions containing a mix of dideoxyribonucleotides which terminate elongation at various points due to the lack of 3' oxygen.
-By randomization, fragments of many different lengths are produce which all indicate the position of a corresponding nucleotide of the dideoxyribonucleotides.
Restriction Fragment Length Polymorphisms
-DNA from different individuals can have variation in restriction sites when treated with restriction enzymes.
-Their analysis is useful for detecting mutations, assessing risk for genetic disorders, and forensic science.
-Vector cloning requires plasmids, restriction enzymes, ligase, and a vector (bacteria).
-"Sticky ends" are single strand sequences at the ends of the cut pieces that matches with other sticky ends made by the same restriction enzyme.
Polymerase Chain Reaction
-Successive cycles of heating and cooling can cause DNA to separate, have primers bind on, elongated by Taq polymerase and the cycle repeats.
-By the end of the third cycle, the first desired sequences are produced. By the end of the 20th cycle, over a million of the desired sequence is produced.
Gel Electrophoresis
-By the end of the third cycle, the first desired sequences are produced. By the end of the 20th cycle, over a million of the desired sequence is produced.
Gel Electrophoresis
-DNA that has been treated by restriction enzymes can be sorted into their respective lengths.
-Smaller fragments move through the gel to the positively-charged terminal (DNA has a negative charge) while larger fragments remain closer to the starting point.
DNA Sequencing
-DNA is replicated in solutions containing a mix of dideoxyribonucleotides which terminate elongation at various points due to the lack of 3' oxygen.
-By randomization, fragments of many different lengths are produce which all indicate the position of a corresponding nucleotide of the dideoxyribonucleotides.
Restriction Fragment Length Polymorphisms
-DNA from different individuals can have variation in restriction sites when treated with restriction enzymes.
-Their analysis is useful for detecting mutations, assessing risk for genetic disorders, and forensic science.
Monday, October 10, 2011
Blog 3: PCR vs. Vector Cloning and PCR vs, DNA Sequencing
PCR vs Vector Cloning
Polymerase Chain Reaction (PCR)
Vector Cloning
Similarities
-PCR and Vector Cloning can produce millions of copies of a desired sequence of DNA or gene in a very short amount of time.
-Gel Electrophoresis is used to isolate the length of DNA containing the gene of interest.
Differences
-PCR requires special heat-resistant taq polymerase obtained from bacteria living in hot springs
-Vector Cloning requires a vector to reproduce the DNA; bacteria such as E.coli.
-The DNA sequence is inserted into the plasmid which is placed back into the bacterium through transformation.
-The DNA sequence is inserted into the plasmid which is placed back into the bacterium through transformation.
PCR vs DNA Sequencing
DNA Sequencing (Sanger's method)
Similarities
-PCR and DNA Sequencing use polymerase in their process for elongation.
-A small known sequence of DNA is required for both processes.
Differences
-DNA Sequencing creates fragments of DNA of various lengths which indicates the position of a nucleotide.
Differences
-DNA Sequencing creates fragments of DNA of various lengths which indicates the position of a nucleotide.
-Special dideoxynucleotides with a fluorescent or radioactive label terminate elongation at random points on the DNA strand to create the various fragment lengths.
-PCR copies the whole template of DNA repeatedly.
Sunday, October 2, 2011
Blog 2: 10 points to know about genetics
[1] DNA and RNA grows from 5'-3'. 5' end contains a free phosphate group attached to deoxyribose while the 3' end contains a free hydroxyl group attached to deoxyribose. New nucleotides can only be added to the 3' end.
[2] Frederick Griffith showed that bacteria rendered harmless could assimilate a foreign substance (DNA) from heat-killed bacteria and become pathogenic. This proved proteins were not the genetic material as heat kills proteins. Griffith called this process transformation.

[3] Watson and Crick's model of semi conservative replication states that DNA molecules produce daughter molecules with one old strand and one newly made strand.

[4] A large team of enzymes and proteins carry out DNA replication at many sites where the DNA has been opened up into a "bubble" with replication forks at the ends.

[6] Transcription synthesizes mRNA from the template strand of a gene. It begins at the promotor region (5' TATA 3') on the template strand and endd when a terminator sequence is transcribed (AAUAA, "Arnold"). RNA polymerase II is used to synthesize the mRNA.
[7] Introns and RNA splicing are responsible for the wide range of proteins a single gene can code for.

[9] On a 5' to 3' mRNA codon, the 3rd base can form pairs with tRNA anti codons that do not follow the Watson and Crick model. This wobble position helps to defend against point mutations (a nucleotide swapped for another). In the case a mutation occurs on the 3rd base, the codon can still be translated to the correct protein (ex. UUC and UUA both code for Phenylalanine)
[10] A gene is a region of DNA whose final product is either a polypeptide or an RNA molecule
[2] Frederick Griffith showed that bacteria rendered harmless could assimilate a foreign substance (DNA) from heat-killed bacteria and become pathogenic. This proved proteins were not the genetic material as heat kills proteins. Griffith called this process transformation.
[3] Watson and Crick's model of semi conservative replication states that DNA molecules produce daughter molecules with one old strand and one newly made strand.
[4] A large team of enzymes and proteins carry out DNA replication at many sites where the DNA has been opened up into a "bubble" with replication forks at the ends.
[5] DNA replication: the leading strand is continuously elongated while the lagging strand is elongated in short pieces called Okazaki fragments.
[6] Transcription synthesizes mRNA from the template strand of a gene. It begins at the promotor region (5' TATA 3') on the template strand and endd when a terminator sequence is transcribed (AAUAA, "Arnold"). RNA polymerase II is used to synthesize the mRNA.
[7] Introns and RNA splicing are responsible for the wide range of proteins a single gene can code for.
[8] Codons are triplets of nucleotides on the mRNA that code for a specific amino acid. 5' AUG 3' starts a polypeptide chain which ends on a stop codon (UGA, UAA, UAG).
[10] A gene is a region of DNA whose final product is either a polypeptide or an RNA molecule
Subscribe to:
Comments (Atom)